|
 |
| Tuberc Respir Dis > Volume 86(3); 2023 > Article |
|
Abstract
Background
Methods
Results
Notes
Authors’ Contributions
Conceptualization: Yeo CD. Methodology: Kwon OB, Han S, Lee HY, Lee SH. Formal analysis: Kim SK, Park CK, Kim JW. Data curation: Kang HS, Kim JS, Kim SJ. Software: Han S. Validation: Han J. Writing - original draft preparation: Kwon OB. Writing - review and editing: Han S, Yeo CD. Approval of final manuscript: all authors.
Fig. 1.

Fig. 2.
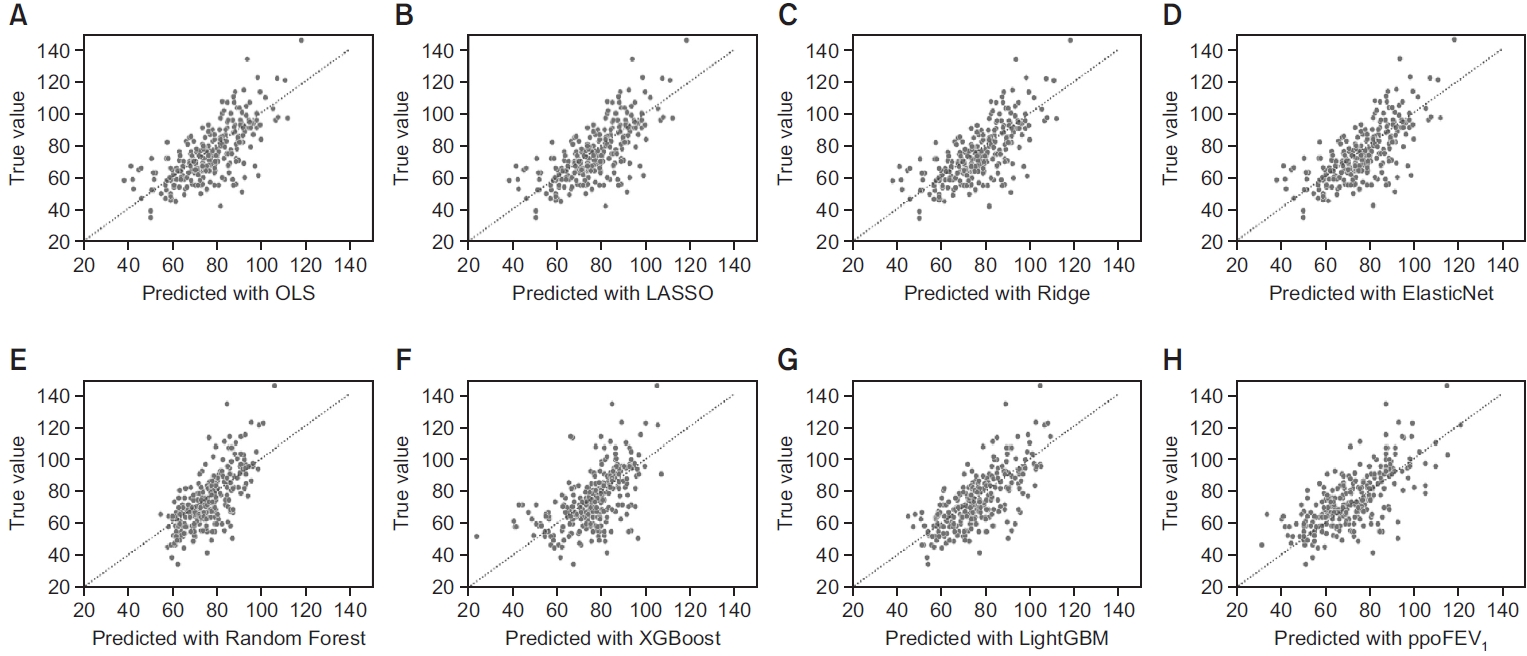
Fig. 3.

Fig. 4.
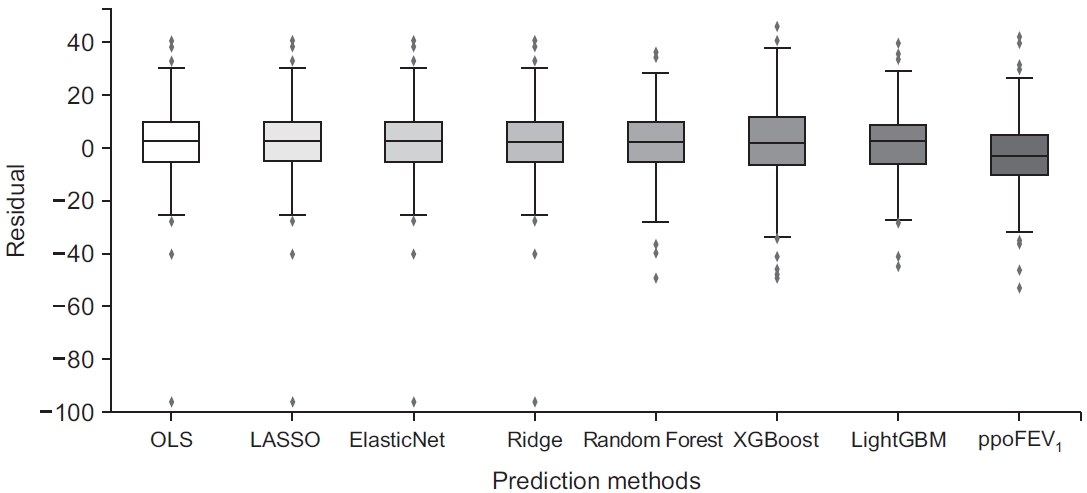
Fig. 5.
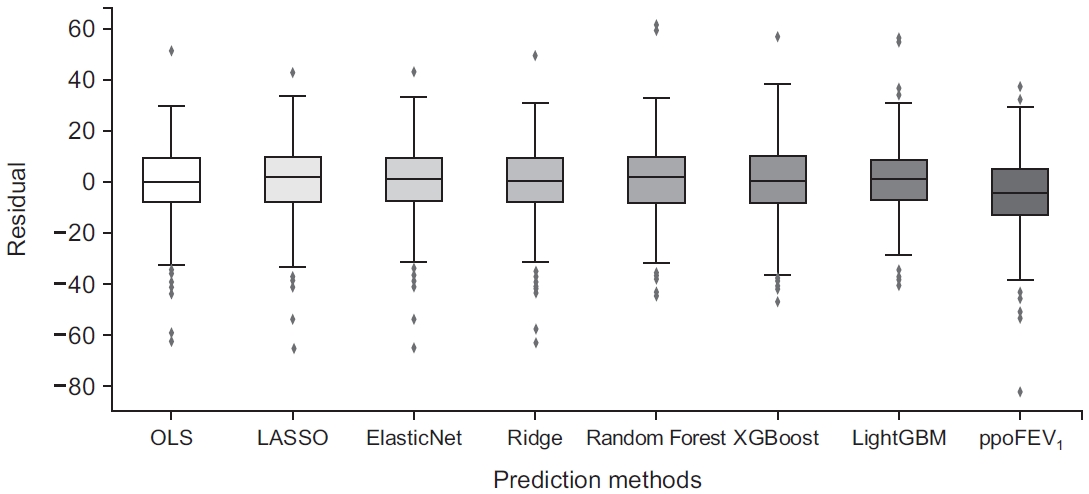
Fig. 6.
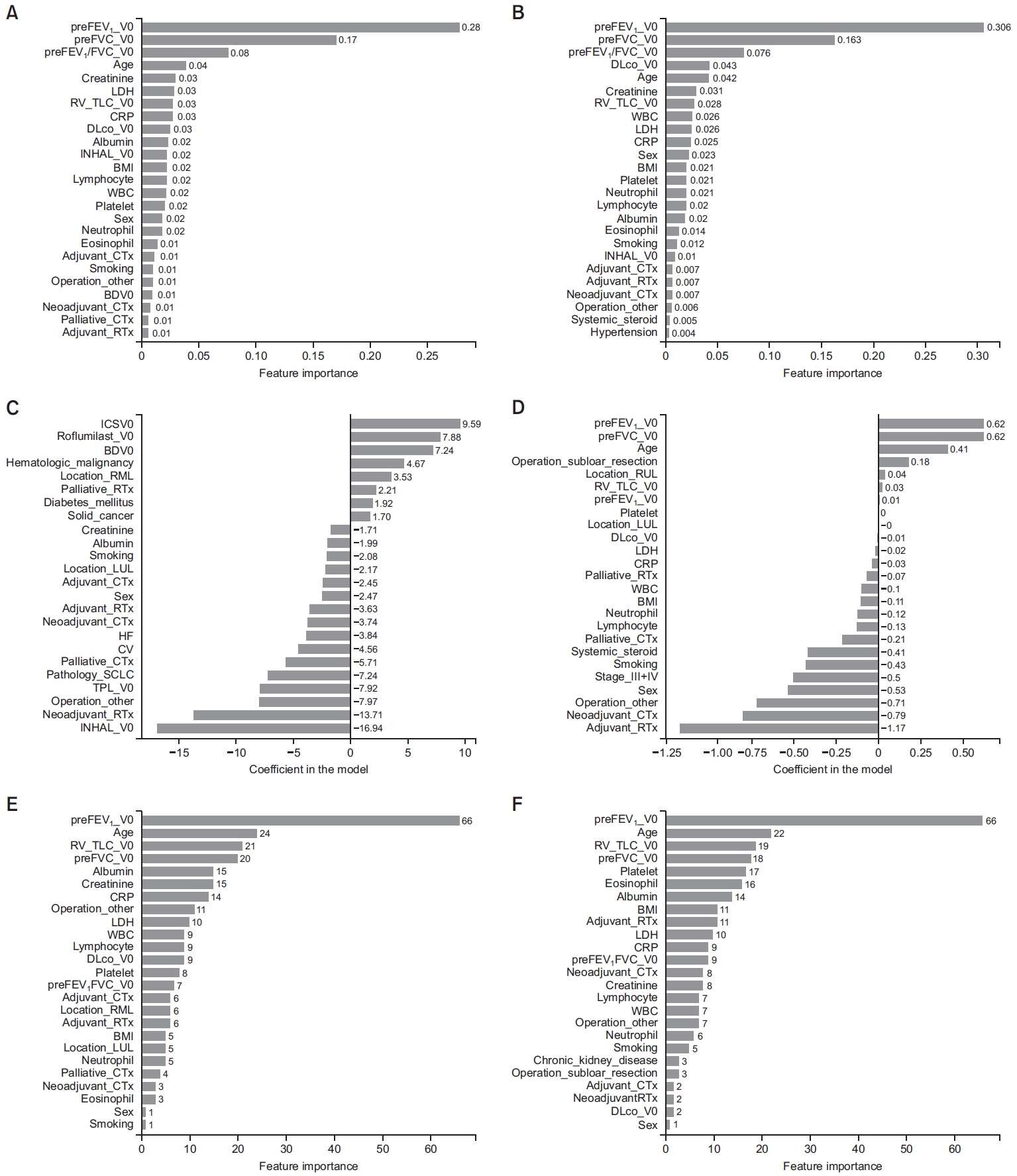
Table 1.
Values are presented as frequency (%) or mean±standard deviation.
SD: standard deviation; BMI: body mass index; SCLC: small cell lung cancer; NSCLC: non-small cell lung cancer; LCC: large cell carcinoma; LCNEC: large cell neuroendocrine carcinoma; RUL: right upper lobe; RML: right middle lobe; RLL: right lower lobe; LUL: left upper lobe; LLL: left lower lobe; IHD: ischemic heart disease; CKD: chronic kidney disease; WBC: white blood cell; CRP: C-reactive protein; LDH: lactate dehydrogenase; V0: time interval from 3 months before surgery to the surgery date; BD: bronchodilator; ICS: inhaled corticosteroid; V1: time interval from 3 to 9 months after the surgery date; FVC: forced vital capacity; FEV1: forced expiratory volume in 1 second; DLCO: diffusing capacity of the lung for carbon monoxide; RV: residual volume; TLC: total capacity; DFS: disease-free survival; OS: overall survival.
Table 2.
β: point estimation; SE: standard error; BMI: body mass index; SCLC: small cell lung cancer; NSCLC: non-small cell lung cancer; RUL: right upper lobe; RML: right middle lobe; RLL: right lower lobe; LUL: left upper lobe; LLL: left lower lobe; WBC: white blood cell; CRP: C-reactive protein; LDH: lactate dehydrogenase; COPD: chronic obstructive pulmonary disease; V0: time interval from 3 months before surgery to the surgery date; BD: bronchodilator; ICS: inhaled corticosteroid; PFT: pulmonary function test; FVC: forced vital capacity; FEV1: forced expiratory volume in 1 second; DLCO: diffusing capacity of the lung for carbon monoxide; RV: residual volume; TLC: total capacity.
Table 3.
GVIF: generalized variation inflation factor; RML: right middle lobe; RUL: right upper lobe; RLL: right lower lobe; LUL: left upper lobe; LLL: left lower lobe; LDH: lactate dehydrogenase; V0: time interval from 3 months before surgery to the surgery date; FVC: forced vital capacity; FEV1: forced expiratory volume in 1 second; DLCO: diffusing capacity of the lung for carbon monoxide.
Table 4.
MSE: mean squared error; ppoFEV1: predicted postoperative forced expiratory volume in 1 second (%) calculated by the formula: ppoFEV1=preFEV1×[1-(number of segments×0.0526)]; OLS: ordinary least squares; LASSO: least absolute shrinkage and selection operator; XGBoost: extreme gradient boosting; LightGBM: light gradient boosting machine; Set II: omitting missing datasets; Set III: imputing missing datasets.
REFERENCES
- TOOLS
-
METRICS

- ORCID iDs
-
Oh Beom Kwon

https://orcid.org/0000-0003-0084-2657Chang Dong Yeo

https://orcid.org/0000-0002-4103-7921 - Funding Information
-
Korean Academy of Tuberculosis and Respiratory Diseases
- Related articles
-
The Prediction of Postoperative Pulmonary Complications in the Elderly Patients.1997 April;44(2)
Prediction of Post-operative Cardiopulmonary function By Perfusion Scan.2001 April;50(4)


 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Data Sharing Statement
Data Sharing Statement Full text via DOI
Full text via DOI Print
Print Download Citation
Download Citation