The Effectiveness of Real-Time PCR Assay, Compared with Microbiologic Results for the Diagnosis of Pulmonary Tuberculosis
Article information
Abstract
Background
The incidence of tuberculosis (TB) in Korea is relatively high compared to the other Organisation for Economic Co-operation and Development (OECD) countries, with a prevalence of 71 per 100,000 in 2012, although the incidence is declining. Real-time polymerase chain reaction (PCR) has been introduced for the rapid diagnosis of TB. Recently, its advantage lies in higher sensitivity and specificity for the diagnosis of TB. This study evaluated the clinical accuracy of real-time PCR using respiratory specimens in a clinical setting.
Methods
Real-time PCR assays using sputum specimens and/or bronchoscopic aspirates from 2,877 subjects were reviewed retrospectively; 2,859 subjects were enrolled. The diagnosis of TB was determined by positive microbiology, pathological findings of TB in the lung and pleura, or clinical suspicion of active TB following anti-TB medication for more than 6 months with a favorable response.
Results
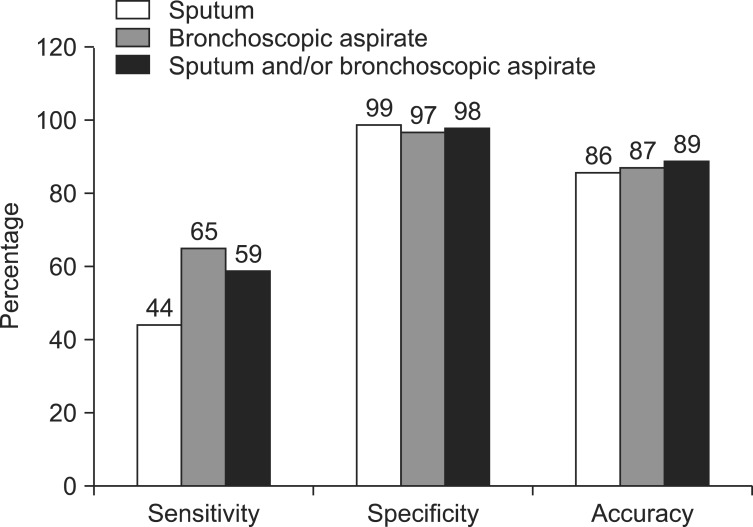
Sensitivity, specificity, and accuracy were 44%, 99%, and 86% from sputum, and 65%, 97%, and 87% from bronchoscopic aspirates, respectively. For overall respiratory specimens, sensitivity was 59%, specificity was 98%, and accuracy increased to 89%.
Conclusion
Positivity in real-time PCR using any respiratory specimens suggests the possibility of active TB in clinically suspected cases, guiding to start anti-TB medication. Real-time PCR from selective bronchoscopic aspirates enhances the diagnostic yield much more when added to sputum examination.
Introduction
The prevalence of tuberculosis (TB) in Korea is declining, but is still high among the Organisation for Economic Co-operation and Development (OECD) countries. Korea had a TB prevalence of 71 per 100,000 in 20121. TB is an important infectious disease, and the Korean government is making an effort to provide rapid diagnosis and treatment through national support and project execution2.
Traditionally, the diagnosis of pulmonary TB is made by bacteriological verification from respiratory specimens. The acid-fast bacilli (AFB) smear test is the gold standard for diagnosis and offers rapid results, but has the disadvantage of low sensitivity and it does not discriminate TB from non-TB mycobacterial infections3,4. AFB culture is a useful method for confirming a diagnosis but can take up to 6 weeks5.
The polymerase chain reaction (PCR) is used widely for early diagnosis of TB6,7,8. Real-time PCR (RT-PCR) has been used recently because it is more rapid than conventional PCR and has excellent reproducibility9,10. However, most previous studies of PCR focused on its accuracy and were limited to proven cases with positive results on AFB smear or culture11,12,13. Our findings emphasized the clinical accuracy of the RT-PCR assay using sputum or bronchoscopic aspiration, in the presence of clinical suspicion of active TB.
Materials and Methods
1. Subjects
Subjects who had RT-PCR performed on sputum and/or bronchoscopic aspirates at the Ewha Womans University Mokdong Hospital, Seoul, Republic of Korea between January 2008 and September 2011 were enrolled. This research was approved by the Institutional Review Board of Ewha Medical Center, Seoul, Republic of Korea (No. 12-11-04). From 2,877 patients taking RT-PCR assay during above period, 2,859 patients were evaluated as the final study population with the exclusion of 18 subjects with positive RT-PCR: 6 subjects due to incomplete medical records and 12 subjects even with anti-TB medication because of inadequacy of 6 months' treatment period. In this group, 888 patients were evaluated human immunodeficiency virus (HIV) infection. Among them, HIV-test was positive in six patients and they had all negative results of RT-PCR. Two patients were treated as active pulmonary TB in that period.
AFB smear and culture were requested for all subjects who were suspected of having active TB in their respiratory system. Their medical records, radiological findings and pathological information were reviewed retrospectively. Active TB was confirmed if one or more of the following criteria were met:
(1) Microbiologic diagnosis based on AFB smear positivity with no evidence of nontuberculous mycobacteria (NTM) or AFB culture positivity.
(2) Histological diagnosis of TB.
(3) Biochemical evidence of TB in pleural fluid based on adenosine deaminase exceeding 50 U/L with lymphocytic predominance and exclusion of any other diseases.
(4) Clinical diagnosis confirmed by the patient taking anti-TB medication for more than 6 months with a favorable response, based on laboratory and radiographic data despite not meeting categories (1)-(3).
We excluded patients who had NTM positive results in RT-PCR regardless of AFB positive result in smear and who had been identified to NTM in AFB culture.
2. AFB smear and culture
The respiratory samples were liquefied and decontaminated by the N-acetyl-L-cysteine 2% sodium hydroxide method14. Following concentration, a part of the sediment from each specimen was used for AFB staining. After using by auramine-rhodamine stain, positive staining specimen was confirmed by Ziehl-Neelsen (ZN) staining. AFB smear was examined using the guidelines of the Centers for Disease Control and Prevention5. Respiratory specimen was inoculated into 3% Ogawa's medium (Eiken Co., Tokyo, Japan). Since July 2008, it was inoculated into a mycobacteria growth indicator tube, BD BBL MGIT 960 (Beckton Dickinson, Sparks, MD, USA) at the same time. Culture using MGIT 960 was performed according to the manufacturer's instructions. MGIT bottles were incubated and monitored for 6 weeks. Positive sample in MGIT system was collected 0.1 mL for confirming AFB by ZN staining and was performed to identify. Three precentage Ogawa's medium was also incubated at 37℃ and examined for growth every week for 8 weeks, until which a final result was reported. Culture positive specimen was identified by MoleculTech MTB-ID v3 (Youngdong Int., Yongin, Korea), which method was to distinguish Mycobacterium tuberculosis (MTB) and MTB-complex by on-tube nested/multiplex PCR. NTM infection was identified by Myco-ID (M&D Inc., Wonju, Korea) using PCR-restriction fragment length polymorphism since 2006. After April 2011, GenoType Mycobacterium CM & AS kit (Hain Lifescience GmbH, Nehren, Germany) was used for identifying NTM subgroup. All methods for identifying infection were performed according to the manufacturer's instructions. Before the start of TB treatment, at least two sputum samples were collected by self-expectoration or by induction with 3% saline. Bronchoscopic aspirates were collected with saline irrigation from the affected part through conventional bronchoscopy.
3. RT-PCR
In the 654 subjects, RT-PCR assays were conducted using both sputum and bronchoscopic aspirate samples. RT-PCR was performed using the AdvanSure TB/NTM RT-PCR kit (LG Life Science, Seoul, Korea) according to the manufacturer's instructions13,15. Three channels were used in the RT-PCR reaction (M. tuberculosis complex, mycobacteria, internal control). Signals for FAM, HEX, and Cy5 (fluorescent material fixed to the TaqMan probe) were measured in each channel. A cycle threshold (Ct) under 35 PCR cycles of rpoB was specific for acid-fast bacteria. M. tuberculosis was considered present if the Ct of rpoB was less than 35 on each signal and greater than or equivalent to that of IS6110. If the Ct of rpoB was less than 35 on each signal, but less than that of IS6110, the results were interpreted as indicative of M. tuberculosis complex with a small number of IS6110 copies or probable co-infection between M. tuberculosis and a non-TB mycobacterium16. A Ct of rpoB less than 35 and Ct of IS6110 over 35 was interpreted as a non-TB mycobacterial infection13.
4. Statistical analysis
Sensitivity, specificity, positive and negative predictive values, and accuracy were calculated based on TB diagnosis data using preselected criteria. The SPSS version 20.0 software (SPSS Inc., Chicago, IL, USA) was used for the statistical analysis.
Results
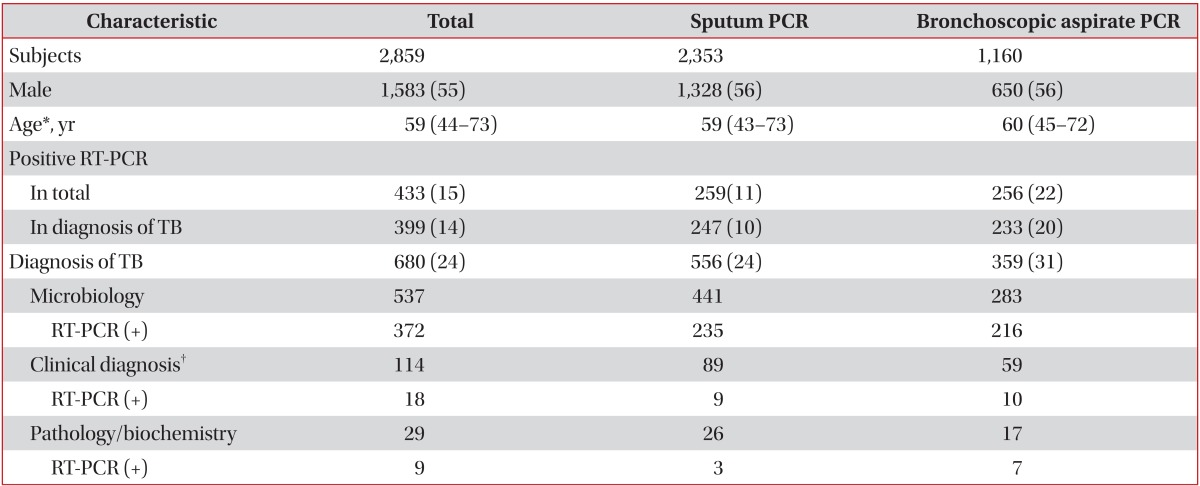
The demographic data for 2,859 subjects are listed in Table 1. The median age was 59 years (interquartile range, 44-73 years), and 55% of the subjects were male. According to the above criteria, 680 subjects (24%) were diagnosed with TB. The diagnosis of active TB among the 680 subjects was based on microbiological positivity in 537 subjects (79%), clinical diagnosis in 114 subjects (17%), and pathology or biochemical data of pleural effusion in 29 subjects (4%). RT-PCR was positive in 22% of the bronchoscopic aspirates, two-fold that from sputum samples (11%). The diagnostic yield of active TB was 24% from sputum samples and 31% from bronchoscopic aspirates. Computed tomography results were available for 81% of the total subjects.
1. RT-PCR of sputum specimens
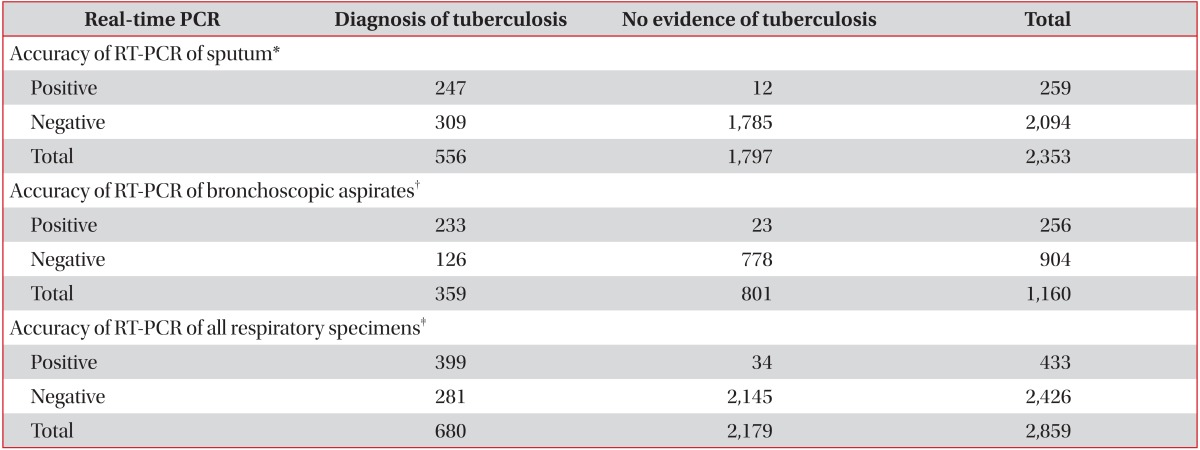
Among the 2,353 subjects whose sputum was tested, 556 were diagnosed with active TB, but only 259 subjects had positive RT-PCR results (Table 1). The RT-PCR assay provided a discrepant diagnosis due to the following: pneumonia infection (28%), active TB (24%), radiologic inactive TB scar (14%), radiological lung mass (11%), bronchiectasis (7%), idiopathic chronic cough (6%), combined pleural effusion (5%), and other medical conditions (5%). Sensitivity of RT-PCR for the prediction of TB infection from sputum specimens was 44% and specificity was 99%, with a positive predictive value of 95%, a negative predictive value of 85%, and an accuracy of 86% (Table 2).
As observation period continued to more than 6 months, 12 subjects were considered false-positives, despite being positive by RT-PCR: nine subjects had negative results in AFB cultures for all respiratory samples and had no evidence of active TB; and three were judged to have inactive TB based on clinical and radiological findings.
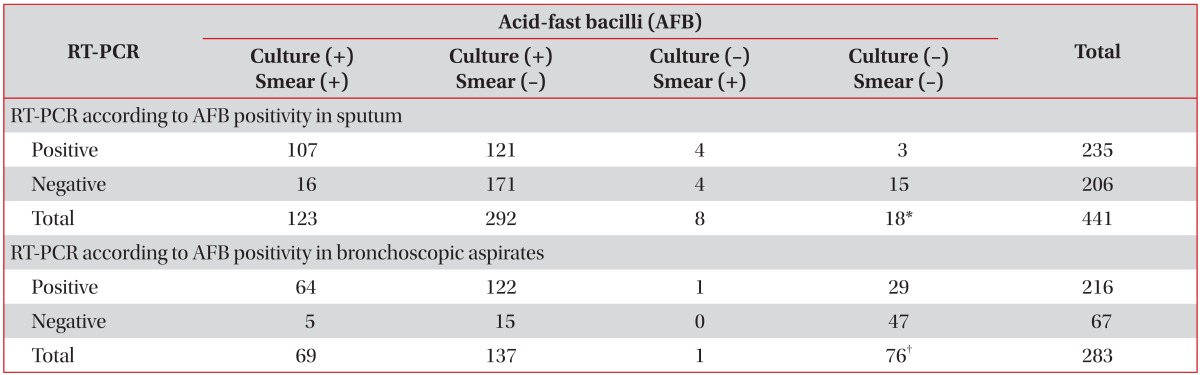
Among the 441 subjects with RT-PCR testing of sputum and microbiological evidence, subgroup analysis was performed in 423 who had positive sputum AFB smear or culture, excluding the 18 cases who had bronchoscopic aspirates positive on either AFB smear or culture. The RT-PCR positivity rate in this subgroup was 55% (232/423). In the 123 subjects whose sputum AFB smear and culture were both positive, the RT-PCR positivity rate was increased to 87% (107/123) (Table 3).
2. RT-PCR of bronchoscopic aspirates
Among the 1,160 subjects whose bronchoscopic aspirates were subjected to RT-PCR, 359 were diagnosed with active TB, and 256 had positive RT-PCR results. The reasons for conducting an RT-PCR assay were as follows: suspicion of active TB (28%), differential diagnosis of pneumonia (27%), radiological lung mass (19%), bronchiectasis (9%), radiological inactive TB scaring (6%), combined pleural effusion (5%), and other medical conditions (6%). The sensitivity of RT-PCR for the prediction of TB infection from bronchoscopic aspirates was 65% and specificity was 97%, for a positive predictive value of 91%, a negative predictive value of 86%, and an accuracy of 87% (Table 2). During over 6 months of observation, 23 subjects were determined to have false-positive results despite being positive by RT-PCR: 13 subjects had negative results in AFB cultures of all respiratory samples and had no evidence of active TB; eight were judged to have inactive TB based on clinical and radiological findings; and two were diagnosed with lung cancer and had negative culture results.
Among the 283 subjects with RT-PCR testing of bronchoscopic aspirates and microbiological evidence, subgroup analysis was performed for 207 who had bronchoscopic aspirates positive on either AFB smear or culture, excluding the 76 cases who were positive on either sputum AFB smear or culture. The rate of RT-PCR positivity in this subgroup was 90% (187/207), which was considerably higher than the 55% in sputum. In the 69 subjects in whom the AFB smear and culture of bronchoscopic aspirates were both positive, the RT-PCR positivity rate was 93% (64/69), slightly higher than that of sputum (Table 3).
3. RT-PCR of all respiratory specimens
Of the 2,859 subjects, the sputum and bronchoscopic samples of 654 were subjected to RT-PCR testing. If one of these samples was positive, the subject was classified as RT-PCR positive. If both results were negative, the subject was classified as RT-PCR negative. The sensitivity of RT-PCR for the prediction of TB infection in any respiratory specimen was 59%, and the specificity was 98%, for a positive predictive value of 92%, a negative predictive value of 88%, and an accuracy of 89% (Table 2). Of the 654 subjects whose two specimens were both subjected to RT-PCR, 82 were positive by RT-PCR in both specimens, and 99% were diagnosed with active TB. A total of 73 subjects were negative by RT-PCR for both specimens, and only 15% were diagnosed with active TB.
4. Comparison of accuracy among specimens
The accuracy of RT-PCR was compared among the samples: sputum, bronchoscopic aspirates, or both. Using both data, the TB detection accuracy was 89% when both samples were used, which was higher than the accuracy of any single specimen (Figure 1).
Discussion
RT-PCR is recommended for any respiratory specimen when TB is suspected clinically, as it has additional benefit in TB detection. Anti-TB medication can be started in a PCR-positive case without waiting for the culture result.
The prevalence of TB is decreasing annually, but it remains an important health issue globally due to its high infectiousness1. Along with the development of molecular biology for rapid diagnosis, several PCR tests for the early detection of M. tuberculosis have been developed since 19908. PCR results are available in a short time and the tests have relatively high sensitivity and specificity, compared to conventional AFB smear or AFB culture. The gene X-pert (Cepheid, Sunnyvale, CA, USA) assay was introduced recently. Its advantage lies in higher sensitivity and specificity for the diagnosis of TB, together with detection of drug resistance17. The relative costs of gene X-pert and RT-PCR differ among countries. RT-PCR has the advantage of detecting non-tuberculous mycobacteria as well as M. tuberculosis.
Most reports have evaluated PCR using known AFB-positive samples17,18,19. In smear-positive specimens, the sensitivity and specificity of PCR are in the range 90%-100%, with a positive predictive value of >95%, whereas in smear-negative specimens, the sensitivity of PCR is reduced to <50%17. In this study, the sensitivity of RT-PCR in AFB smear-positive specimen was observed 89%. Our study differed from previous works in terms of its design. On repeated requests for AFB smear of respiratory specimen, RT-PCR test was sent at one time under the unknown status of smear results, which occurs in clinical practice. The RT-PCR positivity rates in the AFB-positive subjects in both sputum and bronchoscopic aspirates were increased to 55% and 90%, respectively, from 11% and 22% for all subjects, which is comparable with previous reports18,20. Factors that affect RT-PCR sensitivity include the individual effort expended for sputum collection and clinician bias with regard to diagnostic approaches13.
In this study, the RT-PCR specimens were respiratory samples, including bronchoscopic aspirates. Bronchoscopy is an important tool for the evaluation of pulmonary diseases and is useful especially for differential diagnosis between TB and other conditions21,22. Selective aspirates obtained using the bronchoscopic approach is useful in cases in which little sputum can be obtained or in an AFB-smear-negative subject21. The current results suggest that RT-PCR is more sensitive for detection of M. tuberculosis in bronchoscopic aspirates than in sputum. However, when any respiratory sample is RT-PCR positive, it is recommended that anti-TB medication be started in clinically suspected TB. If two specimens are RT-PCR positive, anti-TB medication is strongly recommended regardless of the results of smear and culture. Conversely, in cases in which both specimens are negative, other pulmonary disorders should be considered.
In our study, the false-positive rate was 0.5% in sputum and 2.0% in bronchoscopic aspirates. False-positivity in PCR has been reported to be due to carry-over contamination between specimens, cross-reactions with isolated NTM, or dead tissue debris from previous TB scarring in highly endemic areas13,23,24.
This study had several limitations. It was retrospective in design and included only one institution. In addition, bias according to the indications considered by the clinician when RT-PCR was requested could have affected the results, as mentioned previously.
RT-PCR tests of all available respiratory specimens could increase the TB diagnostic yield further, and RT-PCR of bronchoscopic aspirates is more useful than that of sputum. As various PCR methods for the diagnosis of TB are introduced, verification and comparison is needed. Further, large-scale studies are warranted to confirm our findings.
Acknowledgements
Authors thank Kyung Ae Kong, M.D. for her contribution to the analysis of data.
Notes
No potential conflict of interest relevant to this article was reported.