Recurrent Pseudomonas aeruginosa Infection in Chronic Lung Diseases: Relapse or Reinfection?
Article information
Abstract
Background
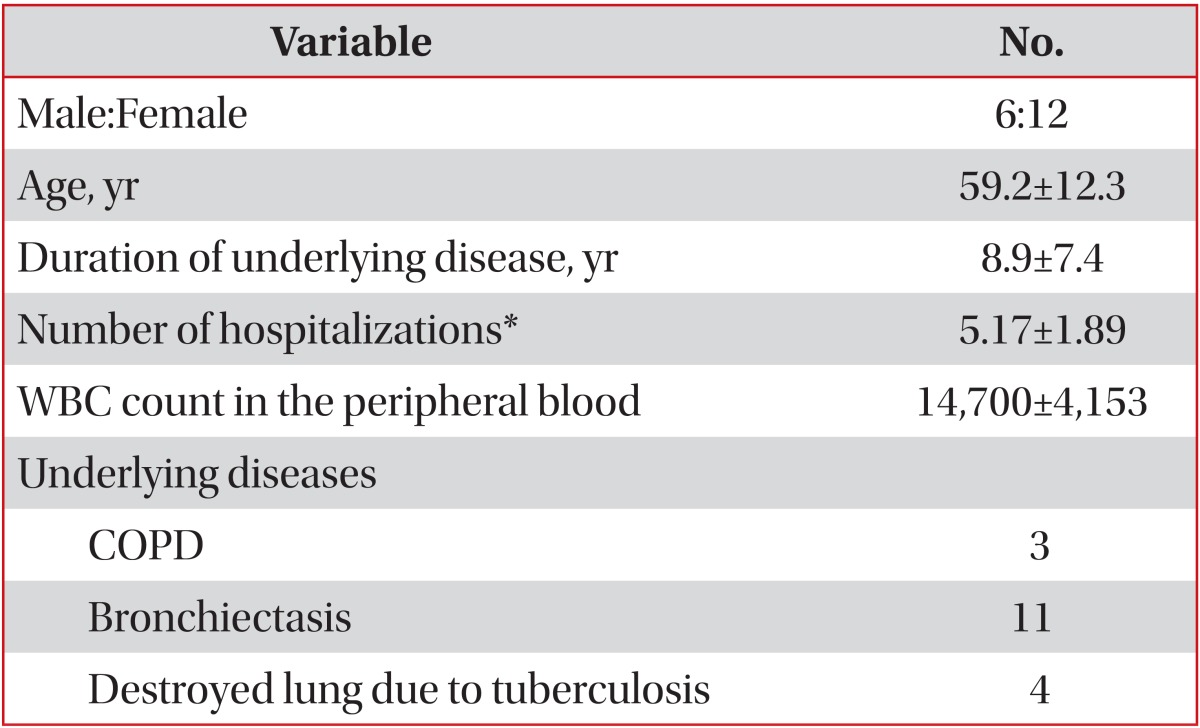
Pseudomonas aeruginosa infection is particularly associated with progressive and ultimately chronic recurrent respiratory infections in chronic obstructive pulmonary disease, bronchiectasis, chronic destroyed lung disease, and cystic fibrosis. Its treatment is also very complex because of drug resistance and recurrence.
Methods
Forty eight cultures from 18 patients with recurrent P. aeruginosa pneumonia from 1998 to 2002 were included in this study. Two or more pairs of sputum cultures were performed during 2 or more different periods of recurrences. The comparison of strains was made according to the phenotypic patterns of antibiotic resistance and chromosomal fingerprinting by pulsed field gel electrophoresis (PFGE) using the genomic DNA of P. aeruginosa from the sputum culture.
Results
Phenotypic patterns of antibiotic resistance of P. aeruginosa were not correlated with their prior antibiotic exposition. Fifteen of 18 patients (83.3%) had recurrent P. aeruginosa pneumonia caused by the strains with same PFGE pattern.
Conclusion
These data suggest that the most of the recurrent P. aeruginosa infections in chronic lung disease occurred due to the relapse of prior infections. Further investigations should be performed for assessing the molecular mechanisms of the persistent colonization and for determining how to eradicate clonal persistence of P. aeruginosa.
Introduction
Pseudomonas aeruginosa is an aerobic gram-negative rod, which does not usually cause infection in the absence of impaired host defenses1. Recurrent P. aeruginosa infection is common in chronic pulmonary diseases like as chronic obstructive lung disease (COPD), bronchiectasis and chronic destroyed lung disease due to tuberculosis in Korea. It is not only resistant to a treatment with antibiotics, but also sometimes fatal in a destructive lung disease due to recurrent infections2,3,4.
Clinically, the same patterns of antibiotic susceptibility test of recurrent pneumonia after a couple of therapy months suggested that the pneumonia caused by P. aeruginosa would be due to predominantly to a reactivation of a previous strain (relapse) rather than to an exogenous reinfection3,4.
Moreover, this pseudomonal infection of lung is notorious to a re-infection or relapse of diseases. Rello et al.5 reported a relapse rate higher as 50% in mechanical ventilated patients with endobronchial intubation or tracheostomy patients in the intensive care unit (ICU). And Talon et al.6 reported that endogenous sources of P. aeruginosa infection are associated with previous infections of patients in the ICU. Therefore, it is very important to determine whether recurrent P. aeruginosa pneumonia is a relapse or re-infection because it provides us with information how long to treat with antibiotics, how to manage the eradication of this troublesome organism and to decide the further research purposes.
Lots of phenotypic markers of P. aeruginosa were used such as pyocin production, pyocin susceptibility, serotyping, antibiotyping and phage susceptibility7. However, there seems to be a poor correlation between in vitro antibiotic susceptibility testing and in vivo antibiotic efficacy. Recently, chromosomal fingerprinting was found to be useful to evaluate epidemiologic pattern8,9. The purpose of this study was to define whether recurrent P. aeruginosa pneumonia was caused by the relapse of previous strains or re-infection of new strains through the molecular typing of P. aeruginosa.
Materials and Methods
Forty-eight episodes of 18 patients with recurrent P. aeruginosa pneumonia were included from 1999 through 2002. Two or more sets of sputum cultures were done during 2 or more different periods of recurrences. Several clinical data were evaluated and analyzed including underlying disease, comorbidities, antibiotic treatment, duration of diseases and susceptibility patterns.
A comparison was made between phenotypic patterns of antibiotic resistance and molecular typing based on pulsed field gel electrophoresis (PFGE) with genomic DNA of P. aeruginosa of sputum cultures.
1. Definition of pneumonia
The diagnosis of pneumonia was considered when new pulmonary infiltrates occurred with at least two of the following criteria: (1) fever>38℃; (2) leukocytosis>10,000 per mm3, and (3) purulent respiratory secretions. Antibiotic treatment of pneumonia was done by the American Thoracic Society (ATS) guideline, 200110. Combination therapy was conducted with two or more anti-pseudomonal antibiotics to which the P. aeruginosa was susceptible in vitro11. At least 2 weeks antibiotic therapy was done after the sputum showed positive culture. The clinical resolution was defined if the patient had complete resolution of all signs and symptoms of pneumonia along with radiologic improvement. Recurrent pneumonia was defined if it occurred after at least two months of complete treatment of the previous pneumonia without an evidence of a new extrapulmonary source of infection.
2. Microbiologic processing
P. aeruginosa strains from sputum cultures were collected and stored at -70℃. Follow-up cultures were performed at each time of pneumonia recurrence. Antimicrobial susceptibility tests were determined using a commercial microdilution broth method, following the manufacturer's recommendations. The considered minimal inhibitory concentration interpretative standards for susceptibility categories were those given by the Korean guideline. Moderately susceptible and resistant bacteria were both regarded as resistant in this study.
3. Pulsed field gel electrophoresis
Genomic DNA for PFGE was prepared as previously reported12,13. Chromosomal DNA plugs were incubated with XbaI. Restriction fragments were separated with an orthogonal field alternation electrophoresis apparatus through 1% agarose gel for a 20 hours run at a constant voltage of 250 V (Figure 1). The pulse time was 20, 10, and 2 seconds for 3, 15, and 2 hours, respectively. PFGE chromosomal similarities of the fragment length patterns were compared between two strains using a dendrogram (Figure 2).
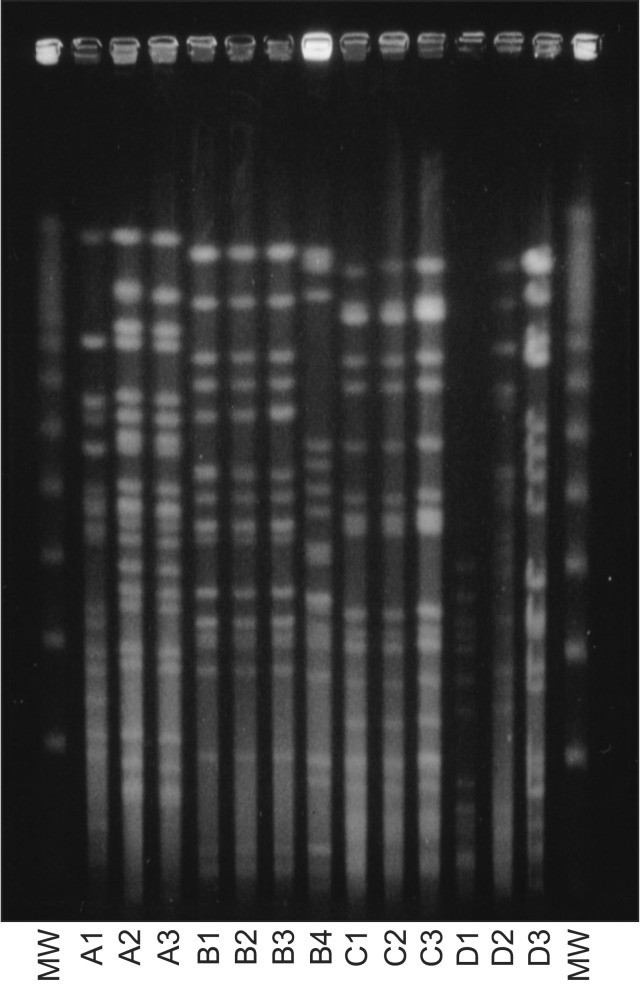
PFGE pattern of Pseudomonas aeruginosa strains isolated from patients with recurrent episodes of pneumonia (MW= molecular weight marker; A1, A2, and A3 are 3 different strains in each pneumonia episode in the same patient). PFGE: pulsed field gel electrophoresis.
4. Statistical analysis
Variables were compared with t-test and one-way analysis of variance (ANOVA).
Results
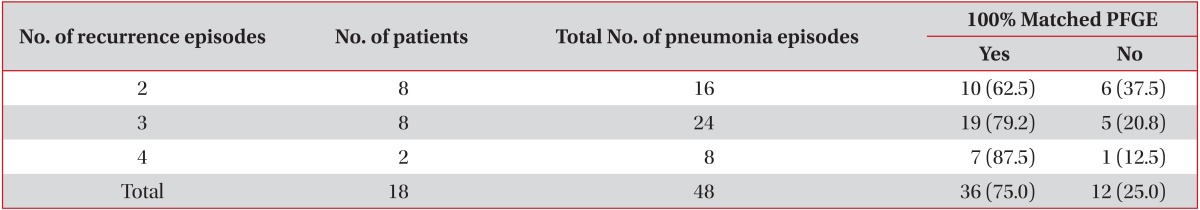
Demographic data and several predisposing factors of recurrent pneumonia were summarized of the included patients (Table 1). The mean duration of underlying disease was 8.9±7.4 years. P. aeruginosa was isolated in 48 episodes of the 18 patients with P. aeruginosa pneumonia. Eight of 18 patients developed a recurrence once; another 8 patients developed a recurrence twice and 2 patients developed a recurrence thrice. Bronchiectasis was the most common underlying disease in recurrent P. aeruginosa pneumonia in this study (11 of 18 patients).
The antibiotic resistance patterns of P. aeruginosa and PFGE patterns were summarized as well as their relationship with prior antibiotic exposure during the first episode of pneumonia (Table 2). Eighteen of 24 paired P. aeruginosa strains showed the same patterns of PFGE. Recurrent P. aeruginosa pneumonia occurred in 75% of the patients (Table 3). Twenty-two of 30 recurrent P. aeruginosa pneumonia cases (73.3%) were caused by the same PFGE patterns. The PFGE patterns were not statistically correlated with the intervals of pneumonia episodes, duration of underlying illness, and prior antibiotic usage.

Antibiotic resistance patterns of Pseudomonas aeruginosa strains and their relationship with prior antibiotic exposition and PFGE patterns

PFGE patterns of Pseudomonas aeruginosa strains isolated from patients with recurrent episodes of pneumonia
Phenotypic patterns of antibiotic resistance of P. aeruginosa were not correlated with their prior antibiotic exposition. Number of episodes of recurrent pneumonia did not affect PFGE patterns statistically (Table 4).
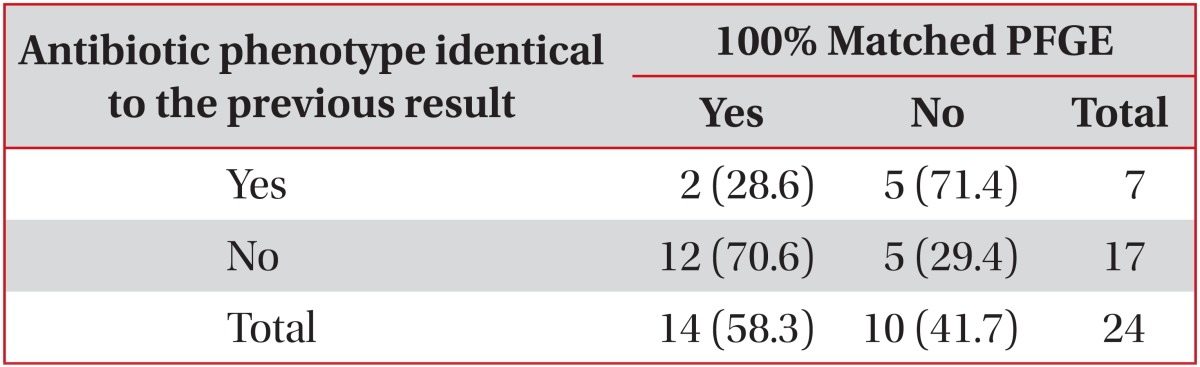
The antibiotic susceptibility profiles of each pneumonia episode were compared with previous resistant patterns (Table 5). Only 7 pneumonic episodes are identical to their previous antibiotic susceptibility profiles. However, PFGE patterns were identical in 75% of pneumonic episodes. PFGE profiles are shown in Figures 1 and 2.
Discussion
P. aeruginosa is one of the most common causes of pneumonia in structural lung diseases such as COPD, bronchiectasis, and destructive lung diseases. Chronic corticosteroid therapy (>10 mg of prednisone per day), broad-spectrum antibiotic therapy for more 7 days in the past month and malnutrition were high risk factors for a pseudomonal infection1.
It is very important to identify whether the resistant strains are recurred or relapsed. Previous studies have reported the incidence of recurrent pneumonia caused by P. aeruginosa in ICU showing a considerable disparity from 3% through 50%7,8.
Various conventional typing systems have been extensively used to study the epidemiology of P. aeruginosa. However, lots of phenotypic markers, such as pyocin production, pyocin susceptibility, serotyping, phage susceptibility, and antibiotyping were poorly correlated with bacterial strains14. And the reproducibility and accuracy of serotypes have been shown to be low. Recently, chromosomal fingerprintings are useful epidemiologic patterns15. Genomic fingerprinting by PFGE was found to be more sensitive than conventional typing methods such as serotyping, pyosin producing and phage typing for the differentiation of P. aeruginosa strains to identify between relapse and reinfection9,10,11. The molecular typing of the different strains isolated from patients with multiple episodes of pneumonia due to P. aeruginosa differentiates between relapse and reinfection.
Our data showed a poor correlation between prior antibiotic usage and PFGE patterns (Tables 2, 3). The number of pneumonia episodes did not affect the PFGE patterns. These data suggest that the most recurrent P. aeruginosa infection in a chronic lung disease occurs due to relapse of the prior infection. The most important mechanism of clinical recurrence is caused by the relapse of prior infected or colonized organisms.
Nevertheless, different molecular genotypes by restriction fragment length polymorphism patterns emerge from variants of the original after antibiotic treatment or are newly acquired strains. Mutations produce new strains that are resistant to anti-pseudomonal antibiotics by mechanisms that include a hyperproduction of chromosomal β-lactamase, an altered DNA gyrase, membrane changes reducing drug accumulation or mechanisms increasing the drug efflux. However, our data suggest that relapse is the most important mechanism of clinical recurrence6.
Niedermann et al.16 showed the persistence of P. aeruginosa in the lower respiratory track of patients with chronic pulmonary diseases for approximately 57% of cultured sputum after antibiotic treatment.
The relapse mechanism is supposed to a persistent colonization in an anatomical destructive bronchial or lung parenchymal focus, such as in bronchiectasis, destroyed lung due to tuberculosis sequele and so forth. And an incomplete or inadequate antibiotic therapy may attribute to a persistent colonization and recurrence as well. Therefore, the control focus should be based on persistent colonization or incomplete eradication of a P. aeruginosa infection.
Typically is P. aeruginosa nonmucoid at its first time of isolation, but after variable periods, often 1 or 2 years, it becomes mucoid strain and produces large amounts of an extracellular polysaccharide called allginate.
Damage to the epithelium, not to the normal epithelium and the structural proteins of lung facilitates the P. aeruginosa adherence. Activated neutrophils spill proteinase enzymes and oxygen radicals in cystic fibrosis or bronchiectatic airways.
Although this study has its limitations which were confirmed as the colonization after recovery of pseudomonal infection, the focus of control should be based on persistent colonization or incomplete eradication of P. aeruginosa infection. These results support the rationale of a long-term treatment with macrolide or fluoroquinolone antibiotics due the anti-inflammatory properties of macrolide antibiotics in a chronic pseudomonal respiratory tract infection.
In conclusion, we should determine whether the occurrence of relapse or re-infection to decide about the antibiotic treatment length and how to permanently eradicate. Also further research purposes should be determined on P. aeruginosa pneumonia in chronic structural destructive lung diseases and further investigation should be done about the molecular mechanism of persistent colonization and how to eradicate the colonial persistence of P. aeruginosa.
Acknowledgements
This work was supported by a Grant from the Inje University, 2003.
Notes
No potential conflict of interest relevant to this article was reported.