|
 |
| Tuberc Respir Dis > Epub ahead of print |
|
Abstract
Background
Methods
Results
Notes
AuthorsŌĆÖ Contributions
Conceptualization: all authors. Methodology: Lee JK, An H, Koh Y. Formal analysis: Lee JK, Lee CH. Data curation: Lee JK, Lee CH. Software: Lee JK, An H, Koh Y. Validation: Lee JK. Investigation: Lee JK, An H. Writing - original draft preparation: Lee JK, Lee CH. Writing - review and editing: all authors. Approval of final manuscript: all authors.
Acknowledgments
Supplementary Material
Supplementary┬ĀTable┬ĀS1.
Supplementary┬ĀTable┬ĀS2.
Supplementary┬ĀTable┬ĀS3.
Supplementary┬ĀTable┬ĀS4.
Supplementary┬ĀTable┬ĀS5.
Supplementary┬ĀTable┬ĀS6.
Supplementary┬ĀTable┬ĀS7.
Fig.┬Ā1.
Table┬Ā1.
| Characteristic | Non-current smoker (n=91) | Current smoker (n=34) | p-value* | COPD GOLD 1 (n=16) | COPD GOLD 2-4 (n=109) | p-valueŌĆĀ |
|---|---|---|---|---|---|---|
| Age, yr | 68.3┬▒8.1 | 67.5┬▒8.4 | 0.296 | 71.1┬▒9.1 | 67.7┬▒7.9 | 0.224 |
| Male sex | 79 (86.8) | 33 (97.1) | 0.096 | 14 (87.5) | 98 (89.9) | 0.769 |
| Body mass index, kg/m2 | 23.8┬▒3.0 | 23.6┬▒3.4 | 0.498 | 23.7┬▒2.4 | 23.7┬▒3.2 | 0.748 |
| Smoking history | <0.001 | 0.555 | ||||
| ŌĆāNever- smoker | 21 (23.1) | 0 | 3 (18.8) | 18 (16.5) | ||
| ŌĆāEx-smoker | 70 (76.9) | 0 | 7 (43.8) | 63 (57.8) | ||
| ŌĆāCurrent smoker | 0 | 34 (100) | 6 (37.5) | 28 (25.7) | ||
| Smoking intensity, pack-yr | 40.5┬▒34.4 | 48.9┬▒18.4 | 0.067 | 39.2┬▒26.6 | 43.3┬▒31.7 | 0.784 |
| SGRQ score | 36.5┬▒16.5 | 35.0┬▒13.2 | 0.681 | 38.5┬▒18.1 | 35.7┬▒15.3 | 0.628 |
| CAT score | 17.2┬▒7.5 | 15.9┬▒7.41 | 0.296 | 18.1┬▒6.8 | 16.7┬▒7.5 | 0.332 |
| mMRC grade | 1.36┬▒0.77 | 1.34┬▒0.68 | 0.971 | 1.31┬▒0.95 | 1.37┬▒0.72 | 0.512 |
| Lung function | ||||||
| ŌĆāFEV1, L | 1.51┬▒0.47 | 1.57┬▒0.46 | 0.428 | 2.14┬▒0.49 | 1.43┬▒0.39 | <0.001 |
| ŌĆāFEV1, % predicted | 61.7┬▒17.9 | 62.5┬▒18.1 | 0.739 | 93.6┬▒10.9 | 57.3┬▒13.5 | <0.001 |
| ŌĆāFVC, L | 3.36┬▒0.82 | 3.43┬▒0.68 | 0.407 | 3.82┬▒0.82 | 3.31┬▒0.76 | 0.006 |
| ŌĆāFVC, % predicted | 92.9┬▒16.7 | 93.1┬▒17.9 | 0.786 | 114.1┬▒16.6 | 89.8┬▒14.7 | <0.001 |
| ŌĆāFEV1/FVC ratio | 0.45┬▒0.12 | 0.46┬▒0.10 | 0.673 | 0.56┬▒0.08 | 0.44┬▒0.11 | <0.001 |
| ŌĆāTLC, % predicted | 115.5┬▒16.2 | 110.1┬▒14.5 | 0.063 | 114.0┬▒15.4 | 114.1┬▒16.0 | 0.790 |
| ŌĆāRV, % predicted | 125.5┬▒36.1 | 115.1┬▒30.8 | 0.199 | 119.8┬▒25.9 | 123.1┬▒36.1 | 0.773 |
| ŌĆāDLCO, % predicted | 80.8┬▒20.7 | 78.5┬▒16.5 | 0.816 | 85.3┬▒18.1 | 79.4┬▒19.8 | 0.153 |
| 6-minute walking distance | 502.6┬▒470.8 | 455.4┬▒83.2 | 0.850 | 470.8┬▒77.6 | 492.5┬▒431.8 | 0.703 |
| Acute exacerbation | ||||||
| ŌĆāMild | 0.37┬▒1.42 | 0.09┬▒0.38 | 0.449 | 0┬▒0 | 0.34┬▒1.31 | 0.186 |
| ŌĆāModerate | 0.84┬▒1.63 | 0.74┬▒1.29 | 0.955 | 1.06┬▒1.34 | 0.77┬▒1.57 | 0.168 |
| ŌĆāSevere | 0.13┬▒0.43 | 0.06┬▒0.24 | 0.463 | 0┬▒0 | 0.13┬▒0.41 | 0.185 |
COPD: chronic obstructive pulmonary disease; GOLD: Global Initiative for Obstructive Diseases; SGRQ: St. GeorgeŌĆÖs Respiratory Questionnaire; CAT: COPD assessment test; mMRC: modified Medical Research Council; FEV1: forced expiratory volume in 1 second; FVC: forced vital capacity; TLC: total lung capacity; RV: residual volume; DLCO: diffusing capacity of the lungs for carbon monoxide.
Table┬Ā2.
| Variable | Non-current smoker (n=91) | Current smoker (n=34) | p-value* | COPD GOLD 1 (n=16) | COPD GOLD 2-4 (n=109) | p-valueŌĆĀ |
|---|---|---|---|---|---|---|
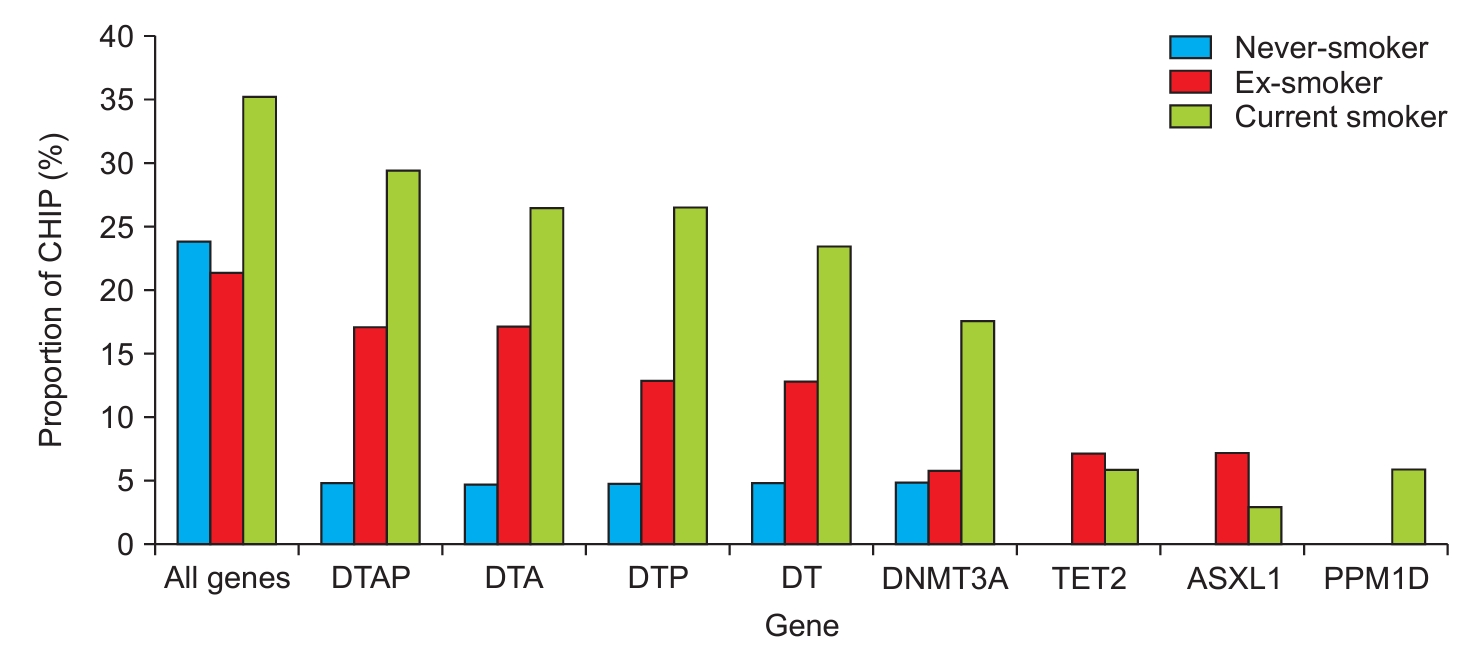
| All genes | 20 (22.0) | 12 (35.3) | 0.131 | 3 (18.8) | 29 (26.6) | 0.503 |
| DTAP | 13 (14.3) | 10 (29.4) | 0.053 | 2 (12.5) | 21 (19.3) | 0.516 |
| DTA | 13 (14.3) | 9 (26.5) | 0.113 | 1 (6.3) | 21 (19.3) | 0.204 |
| DTP | 10 (11.0) | 9 (26.5) | 0.033 | 1 (6.3) | 18 (16.5) | 0.288 |
| DT | 10 (11.0) | 8 (23.5) | 0.077 | 0 | 18 (16.5) | 0.080 |
| DNMT3A | 5 (5.5) | 6 (17.6) | 0.034 | 0 | 11 (10.1) | 0.185 |
| TET2 | 5 (5.5) | 2 (5.9) | 0.933 | 0 | 7 (6.4) | 0.299 |
| ASXL1 | 5 (5.5) | 1 (2.9) | 0.554 | 1 (6.3) | 5 (4.6) | 0.772 |
| PPM1D | 0 | 2 (5.9) | 0.020 | 1 (6.3) | 1 (0.9) | 0.114 |
CHIP: clonal hematopoiesis of indeterminate potential; COPD: chronic obstructive pulmonary disease; GOLD: Global Initiative for Obstructive Diseases; DTAP: combination of DNMT3A, TET2, ASXL1, and PPM1D; DTA: combination of DNMT3A, TET2, and ASXL1; DTP: combination of DNMT3A, TET2, and PPM1D; DT: combination of DNMT3A and TET2; DNMT3A: DNA methyltransferase 3 alpha; TET2: tet methylcytosine dioxygenase 2; ASXL1: ASXL transcriptional regulator 1; PPM1D: protein phosphatase, Mg2+/Mn2+ dependent 1D.
Table┬Ā3.
| Variable |
Univariable |
Multivariable |
||
|---|---|---|---|---|
| OR (95% CI) | p-value | aOR (95% CI)* | p-value | |
| All genes | 1.94 (0.82ŌłÆ4.58) | 0.132 | 2.04 (0.84ŌłÆ4.94) | 0.115 |
| DTAP | 2.50 (0.97ŌłÆ6.42) | 0.057 | 2.41 (0.92ŌłÆ6.31) | 0.072 |
| DTA | 2.16 (0.83ŌłÆ5.65) | 0.117 | 2.06 (0.78ŌłÆ5.47) | 0.147 |
| DTP | 2.92 (1.07ŌłÆ7.97) | 0.037 | 2.80 (1.01ŌłÆ7.79) | 0.048 |
| DT | 2.49 (0.89ŌłÆ6.98) | 0.082 | 2.37 (0.83ŌłÆ6.74) | 0.106 |
| DNMT3A | 3.69 (1.04ŌłÆ13.0) | 0.043 | 4.03 (1.09ŌłÆ14.0) | 0.037 |
| TET2 | 1.08 (0.20ŌłÆ5.82) | 0.933 | 0.89 (0.16ŌłÆ4.88) | 0.890 |
| ASXL1 | 0.52 (0.06ŌłÆ4.63) | 0.559 | 0.49 (0.06ŌłÆ4.40) | 0.524 |
| PPM1D | - | - | - | - |
CHIP: clonal hematopoiesis of indeterminate potential; OR: odds ratio; CI: confidence interval; aOR: adjusted odds ratio; DTAP: combination of DNMT3A, TET2, ASXL1, and PPM1D; DTA: combination of DNMT3A, TET2, and ASXL1; DTP: combination of DNMT3A, TET2, and PPM1D; DT: combination of DNMT3A and TET2; DNMT3A: DNA methyltransferase 3 alpha; TET2: tet methylcytosine dioxygenase 2; ASXL1: ASXL transcriptional regulator 1; PPM1D: protein phosphatase, Mg2+/Mn2+ dependent 1D.
Table┬Ā4.
| Variable |
Univariable |
Multivariable |
||
|---|---|---|---|---|
| OR (95% CI) | p-value | aOR (95% CI)* | p-value | |
| All genes | 1.57 (0.42ŌłÆ5.91) | 0.504 | 1.99 (0.50ŌłÆ8.02) | 0.332 |
| DTAP | 1.67 (0.35ŌłÆ7.92) | 0.518 | 2.33 (0.44ŌłÆ12.4) | 0.324 |
| DTA | 3.58 (0.45ŌłÆ28.6) | 0.229 | 4.80 (0.54ŌłÆ42.4) | 0.158 |
| DTP | 2.97 (0.37ŌłÆ23.9) | 0.307 | 4.12 (0.45ŌłÆ37.6) | 0.210 |
| DT | - | - | - | - |
| DNMT3A | - | - | - | - |
| TET2 | - | - | - | - |
| ASXL1 | 0.72 (0.08ŌłÆ6.60) | 0.772 | 0.86 (0.09ŌłÆ8.49) | 0.896 |
| PPM1D | 0.14 (0.01ŌłÆ2.34) | 0.171 | 0.25 (0.01ŌłÆ5.15) | 0.372 |
CHIP: clonal hematopoiesis of indeterminate potential; COPD: chronic obstructive pulmonary disease; OR: odds ratio; CI, confidence interval; aOR, adjusted odds ratio; DTAP: combination of DNMT3A, TET2, ASXL1, and PPM1D; DTA: combination of DNMT3A, TET2, and ASXL1; DTP: combination of DNMT3A, TET2, and PPM1D; DT: combination of DNMT3A and TET2; DNMT3A: DNA methyltransferase 3 alpha; TET2: tet methylcytosine dioxygenase 2; ASXL1: ASXL transcriptional regulator 1; PPM1D: protein phosphatase, Mg2+/Mn2+ dependent 1D.
Table┬Ā5.
All analysis were adjusted by age, sex, and smoking intensity.
CHIP: clonal hematopoiesis of indeterminate potential; SE: standard error; aOR: adjusted odds ratio; CI: confidence interval; DTAP: combination of DNMT3A, TET2, ASXL1, and PPM1D; DTA: combination of DNMT3A, TET2, and ASXL1; DTP: combination of DNMT3A, TET2, and PPM1D; DT: combination of DNMT3A and TET2; DNMT3A: DNA methyltransferase 3 alpha; TET2: tet methylcytosine dioxygenase 2; ASXL1: ASXL transcriptional regulator 1; PPM1D: protein phosphatase, Mg2+/Mn2+ dependent 1D.
References
- TOOLS
-
METRICS

-
- 0 Crossref
- Scopus
- 568 View
- 27 Download
- ORCID iDs
-
Jung-Kyu Lee

https://orcid.org/0000-0001-5060-7255Chang-Hoon Lee

https://orcid.org/0000-0001-9960-1524 - Related articles


 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Data Sharing Statement
Data Sharing Statement Full text via DOI
Full text via DOI Supplement1
Supplement1 Print
Print Download Citation
Download Citation